This page is not compatible with Internet Explorer.
For security reasons, we recommend that you use an up-to-date browser, such as Microsoft Edge, Google Chrome, Safari, or Mozilla Firefox.
Molecular Diffusion Simulation
with VGSTUDIO MAX
Simulate molecular diffusion on CT scans of different materials using the Transport Phenomena Simulation Module for VGSTUDIO MAX.
Simulation of Molecular Diffusion
Diffusion describes the flux of molecules from a region of higher concentration to one of lower concentration.
The Transport Phenomena Simulation Module for VGSTUDIO MAX:
- Computes the stationary molecular concentration and flux fields, which are reached asymptotically when a sample is connected to reservoirs of different molar concentrations, whereby the pores of the sample are assumed to be completely filled with the solvent and the concentration in the reservoirs is assumed to be constant, like in very large reservoirs.
- Works directly on voxel data and makes use of the sub-voxel accurate, local adaptive surface determination in VGSTUDIO MAX.
- Has "experiment mode" for performing a virtual experiment for the diffusion of molecules and "tensor mode" for calculating the molecular diffusivity tensor.
The molecular diffusion module is based on following differential equations for stationary concentration and molar flux fields in a two-component material:

ₐ
where Ω is the entire simulation domain and Ωₐ is the domain of component a (with a = 1, 2). It is assumed that Ω₁ and Ω₂ do not overlap and their union equals Ω. C is the molecular concentration, J is the molar flux, Dₐ is the diffusion coefficient of component a, Δ is the Laplace operator, and grad is the gradient operator.
Experiment Mode
In experiment mode, the software performs a virtual experiment on the CT data of a structure, simulating the transport of molecules through the structure from an inlet plane towards an outlet plane parallel to each other. Perpendicular to the inlet and outlet plane, sealed or embedded boundary conditions can be defined. A concentration difference must be specified as driving quantity for the flux.
Results of Calculation
The results are provided as 2D and 3D visualizations of molar flux as well as molar concentration. Molecular diffusion directions can be visualized by streamlines in 2D and 3D:
- Molar flux
- Molar concentration
- Streamlines of molecular diffusion
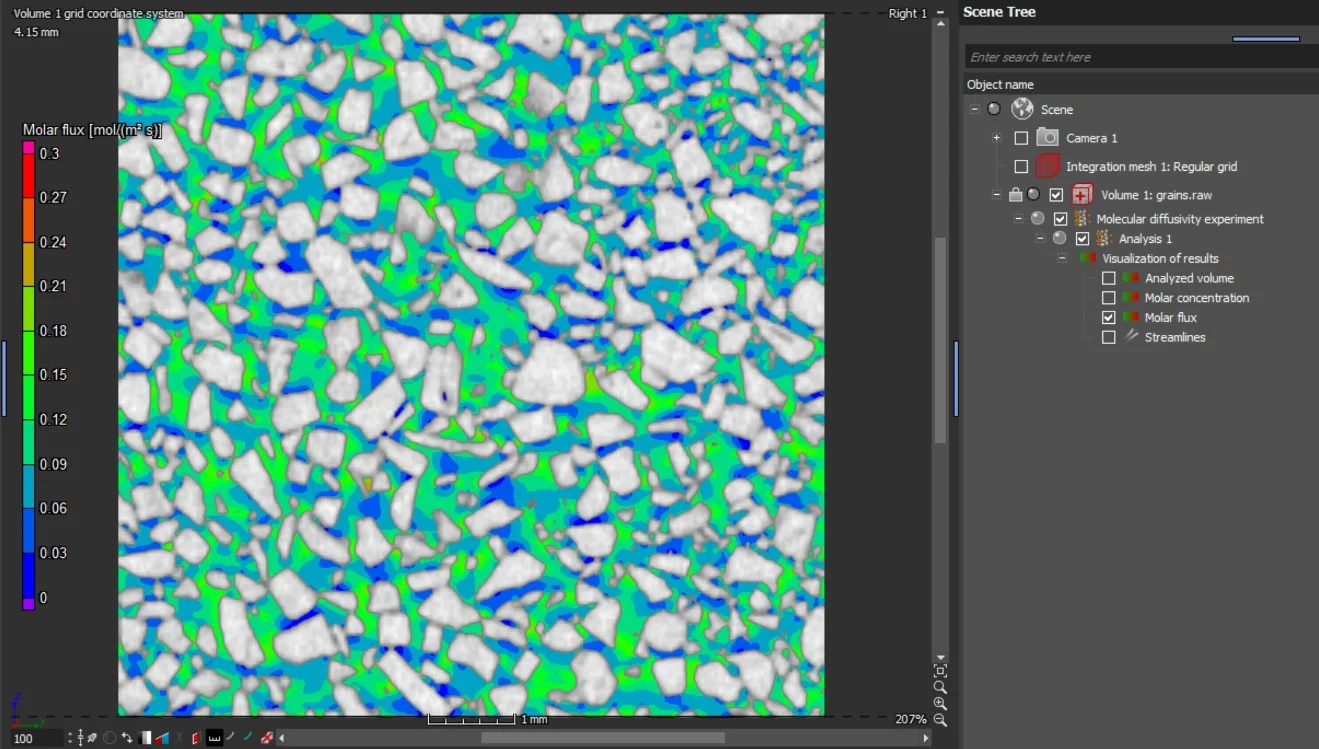
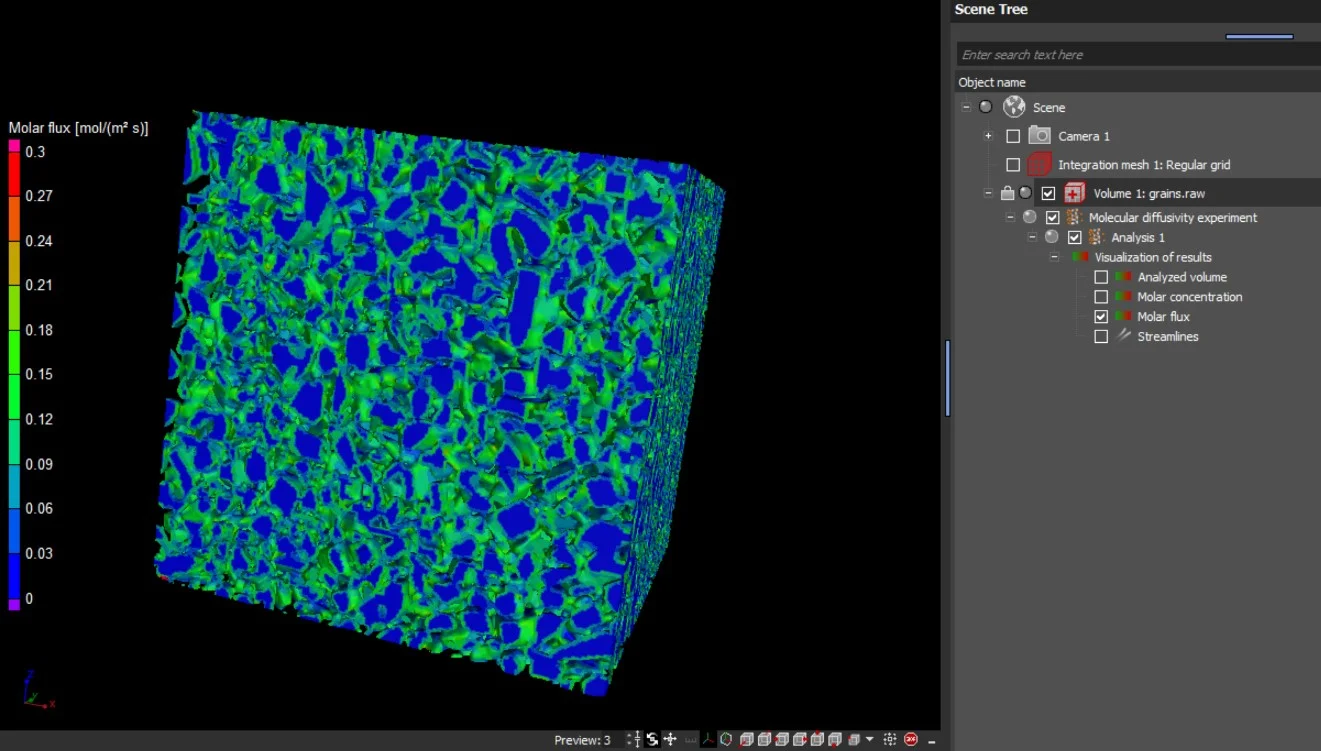
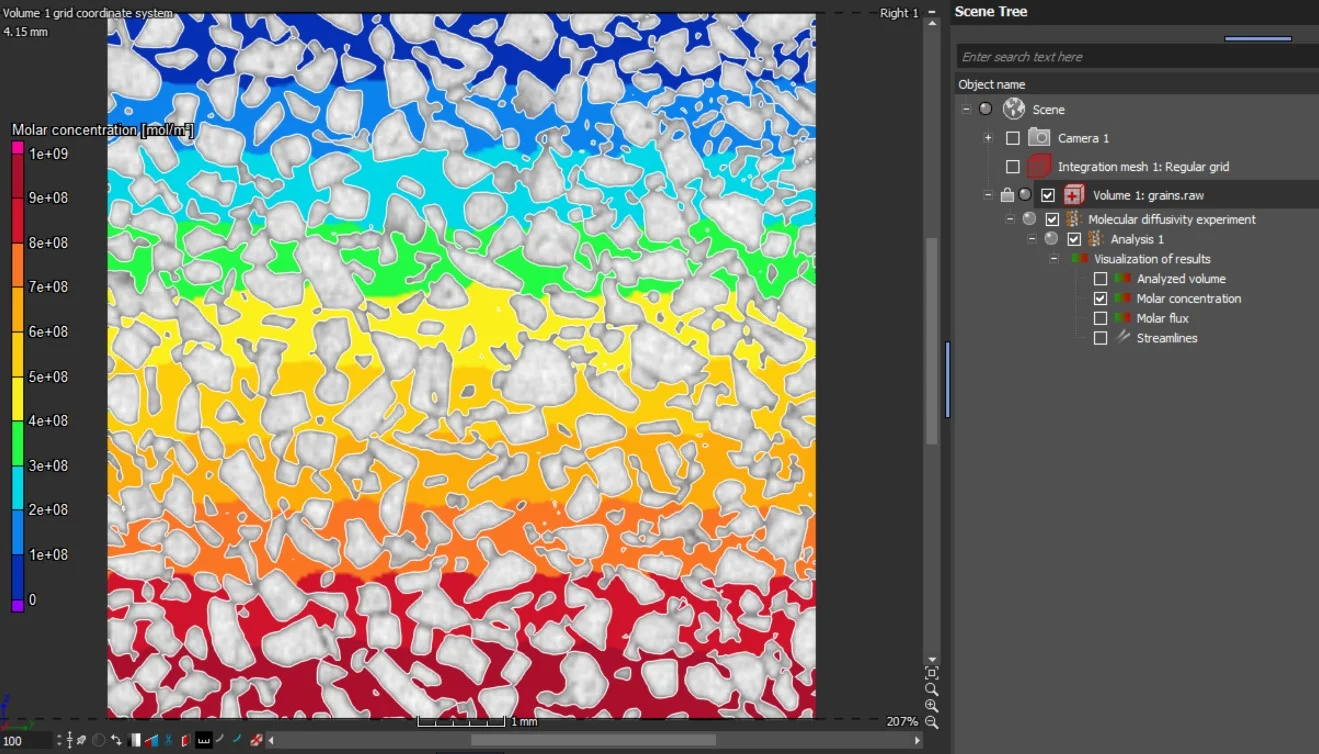
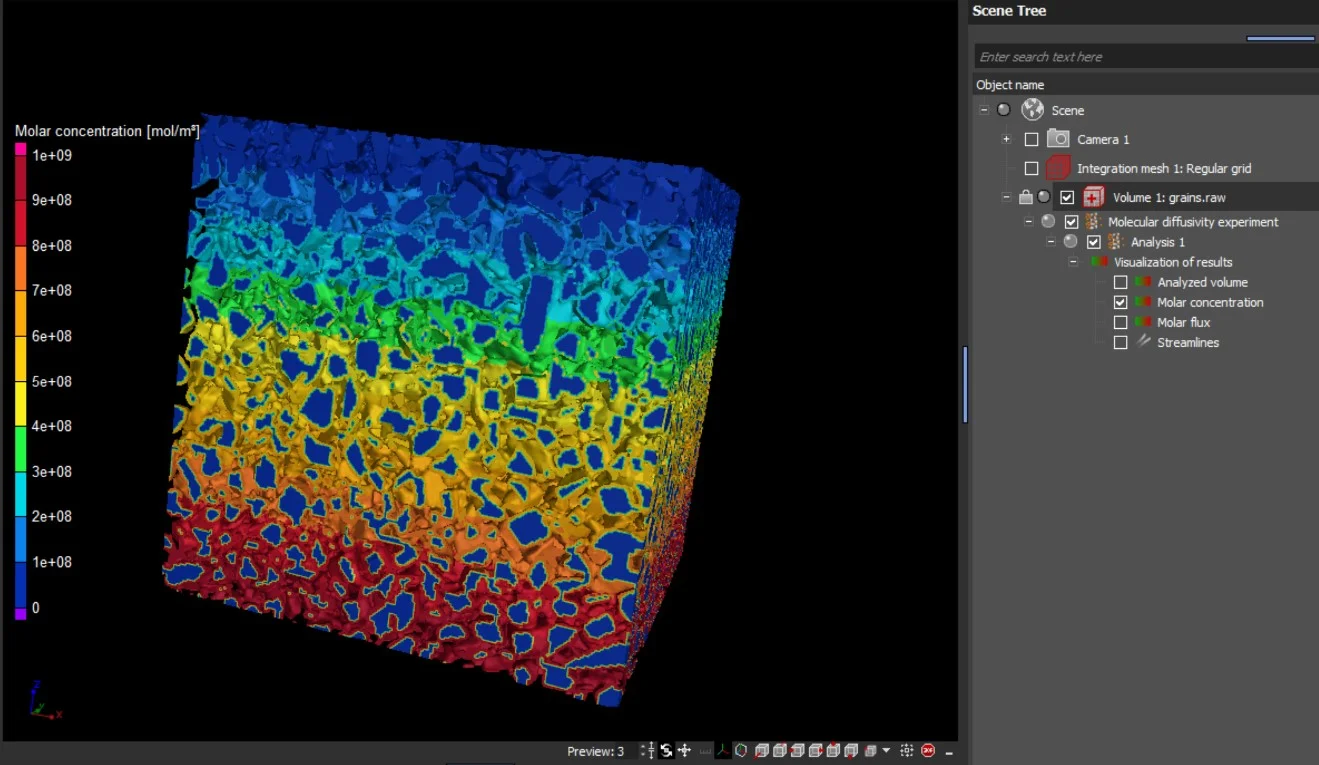
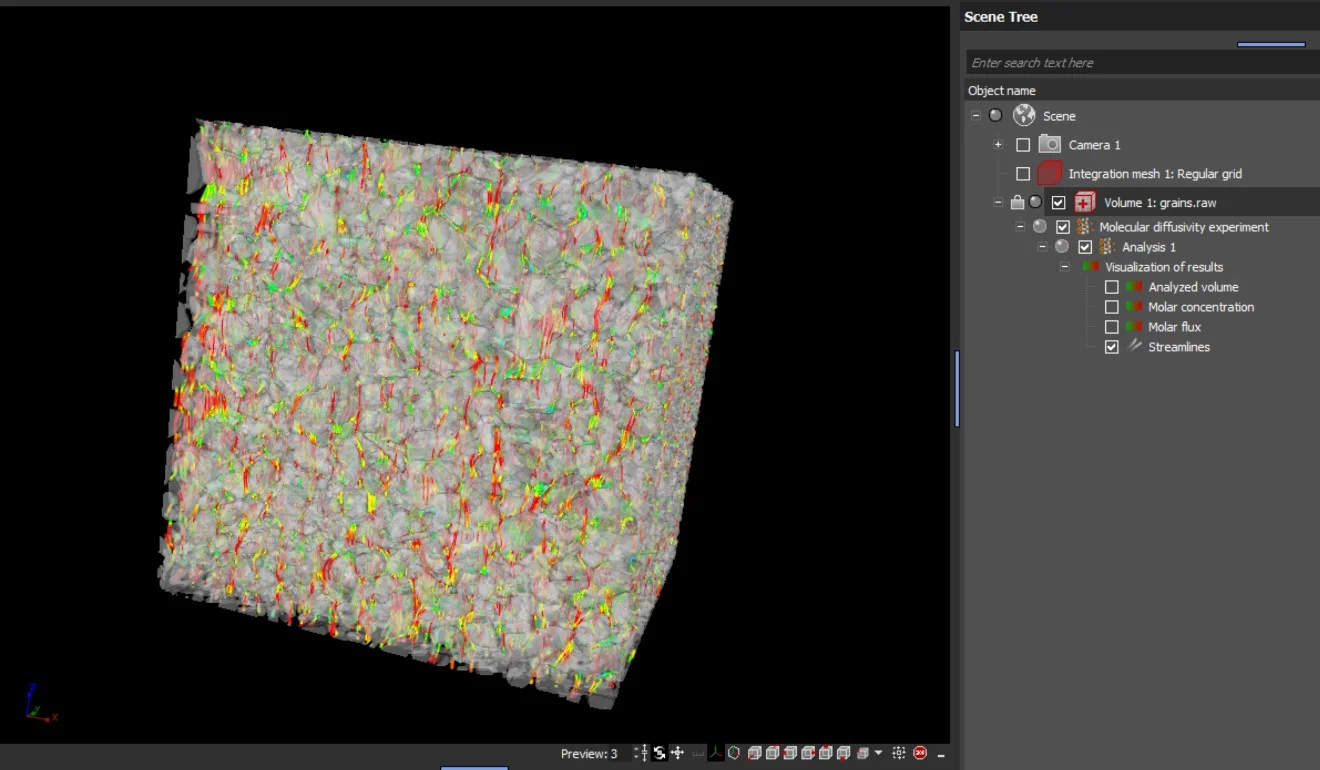
Additional Results
The following additional results are provided in a table view:
- Effective diffusion coefficient
- Diffusive tortuosity (geometric and algebraic)
- Formation factor
- Void fraction
- Total mass transport
- Concentration gradient
The following results can be viewed as curve plots along the flow direction:
- Molar flux
- Void fraction
- Total mass transport
Tensor Mode
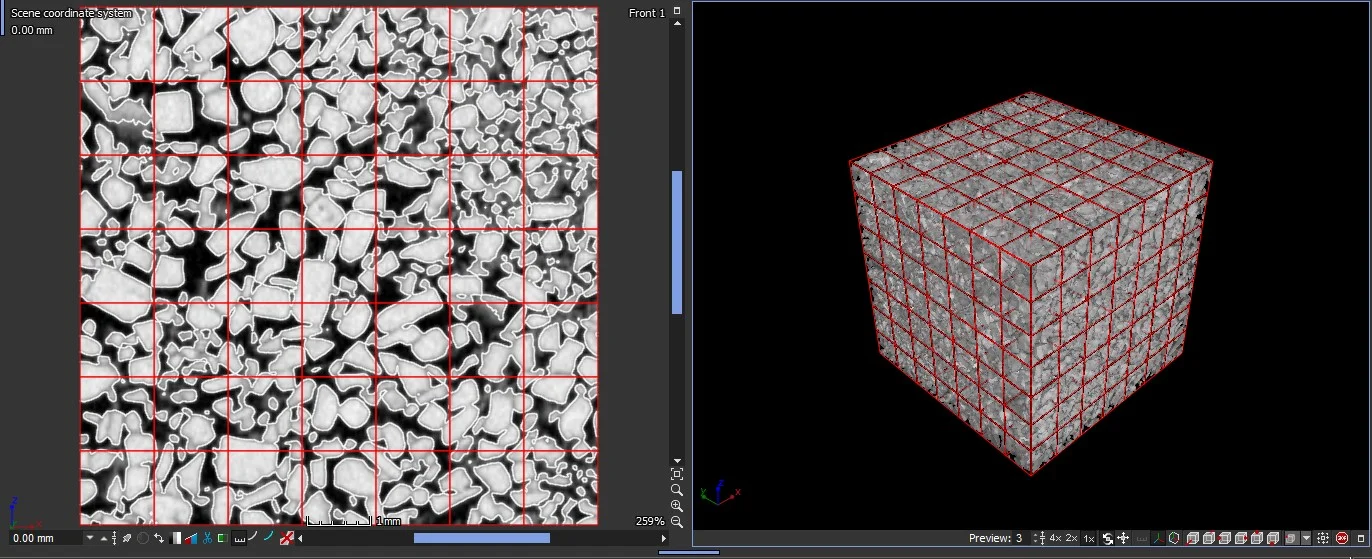
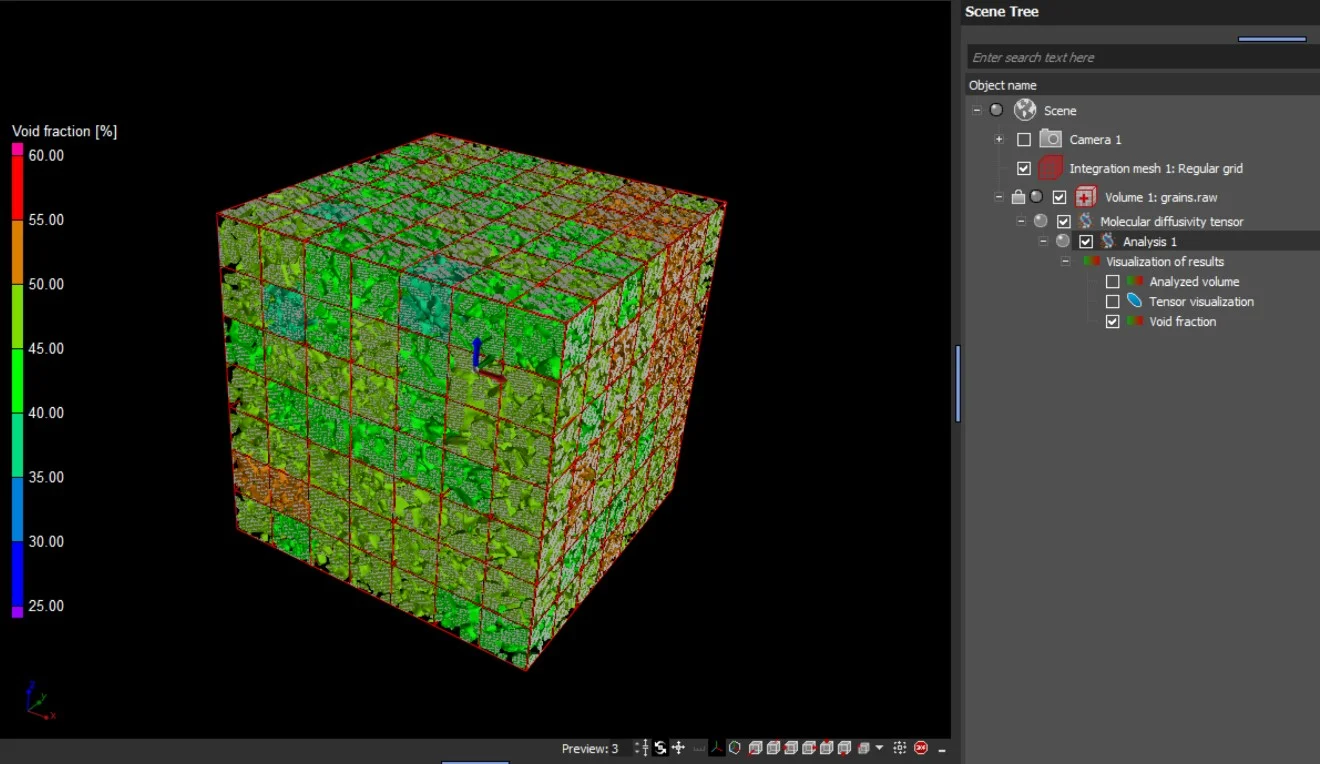
In tensor mode, the software calculates components of the molecular diffusity tensor. The calculation of the molecular diffusity tensor can be done on the whole structure or for increments of the structure by means of an integration mesh.
Results
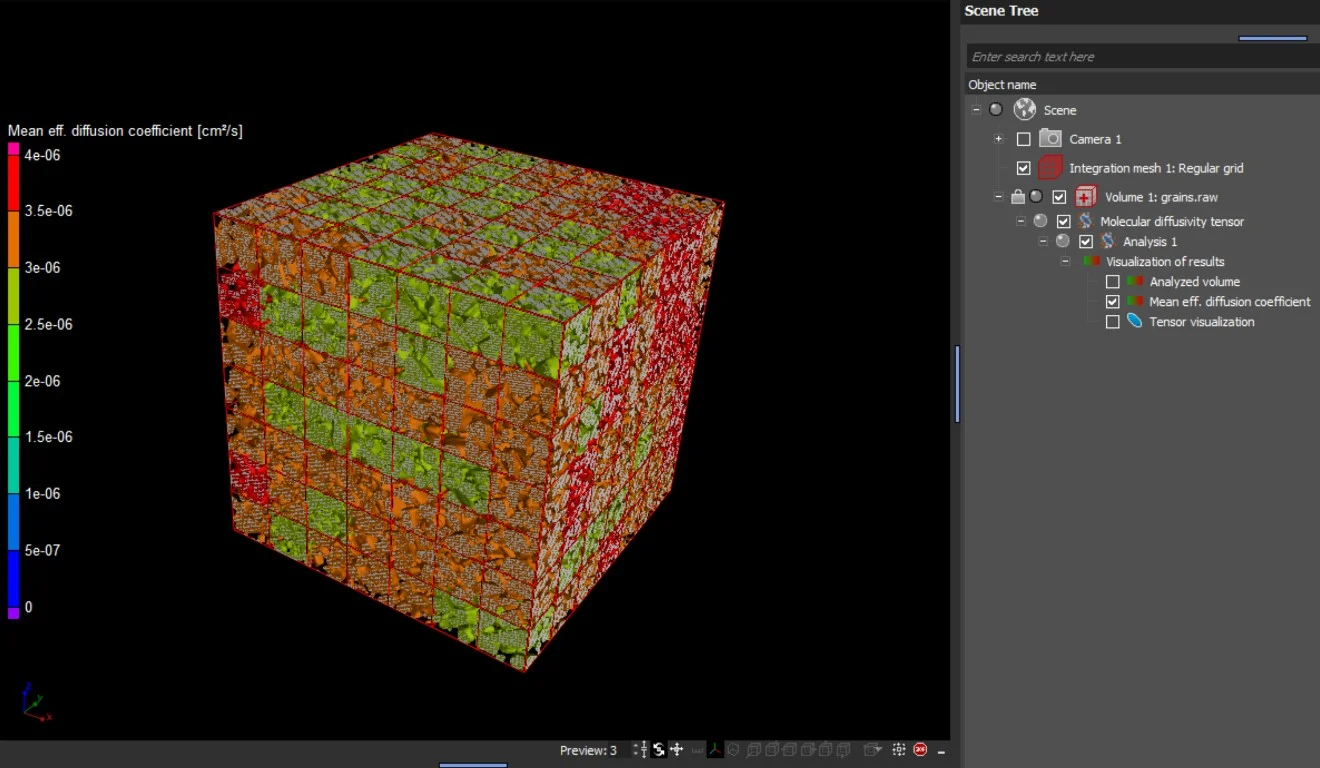
The software provides you molecular diffusion tensor results as 2D and 3D visualizations.
- Results are displayed as an ellipsoid in the center of the simulated volume.
- When the analysis is performed on an integration mesh, results are displayed as an ellipsoid in the center of each cell.
- The main axes of the ellipsoid are aligned with the eigenvectors of the tensor while the lengths of the axes are proportional to the eigenvalues of the tensor.
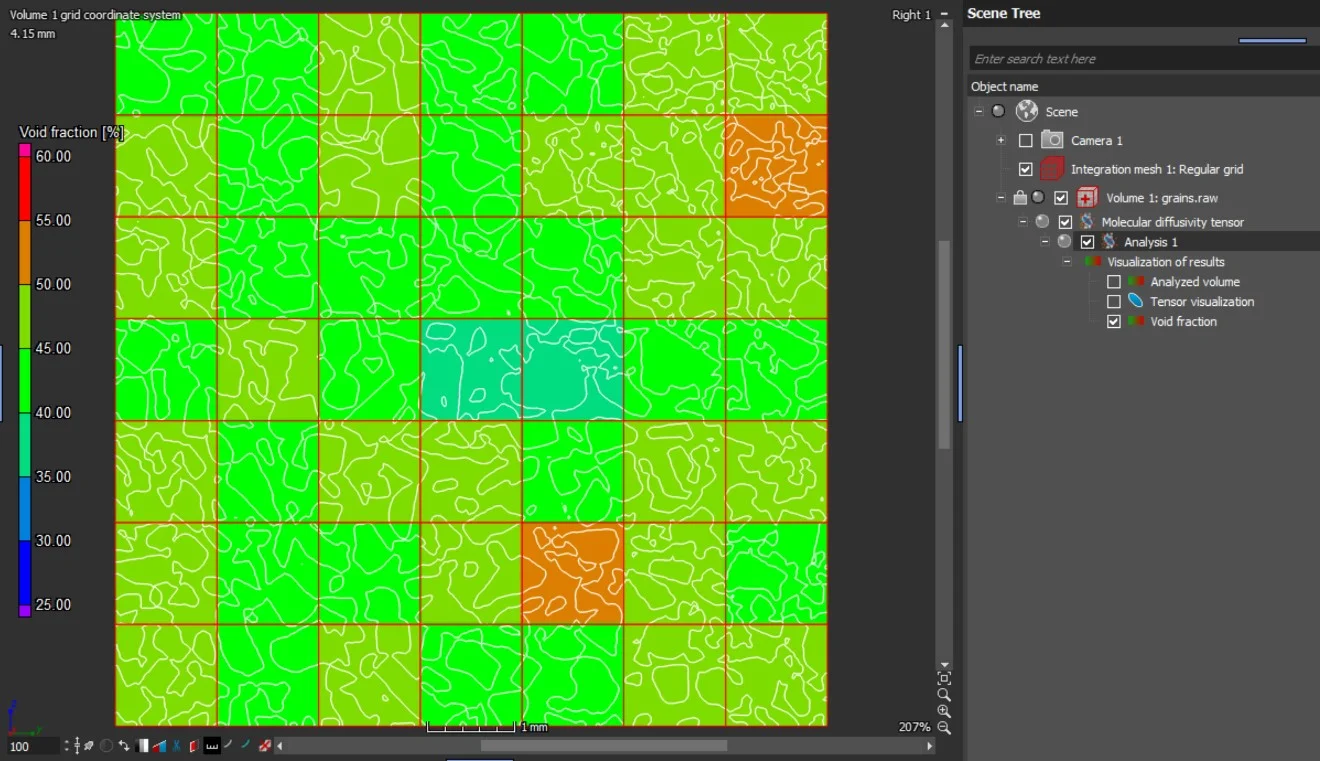
- When the analysis is performed on an integration mesh, the mean effective molecular diffusion tensor and the void fraction, as well as the eigenvalues of the molecular diffusion tensor, can also be visualized in 2D and 3D.


In addition to the tensors' eigenvalues and eigenvectors, the components of the effective diffusion coefficient tensor with respect to the simulation coordinate system are provided in a table view.
Benefits
Easy to use
- No simulation expertise required
- No meshing required
Efficient
- Seamless workflow from material segmentation to simulation in one software
Realistic
- All microstructural details are captured by a subvoxel accurate surface determination